Protein damage
Protein damage susceptibility
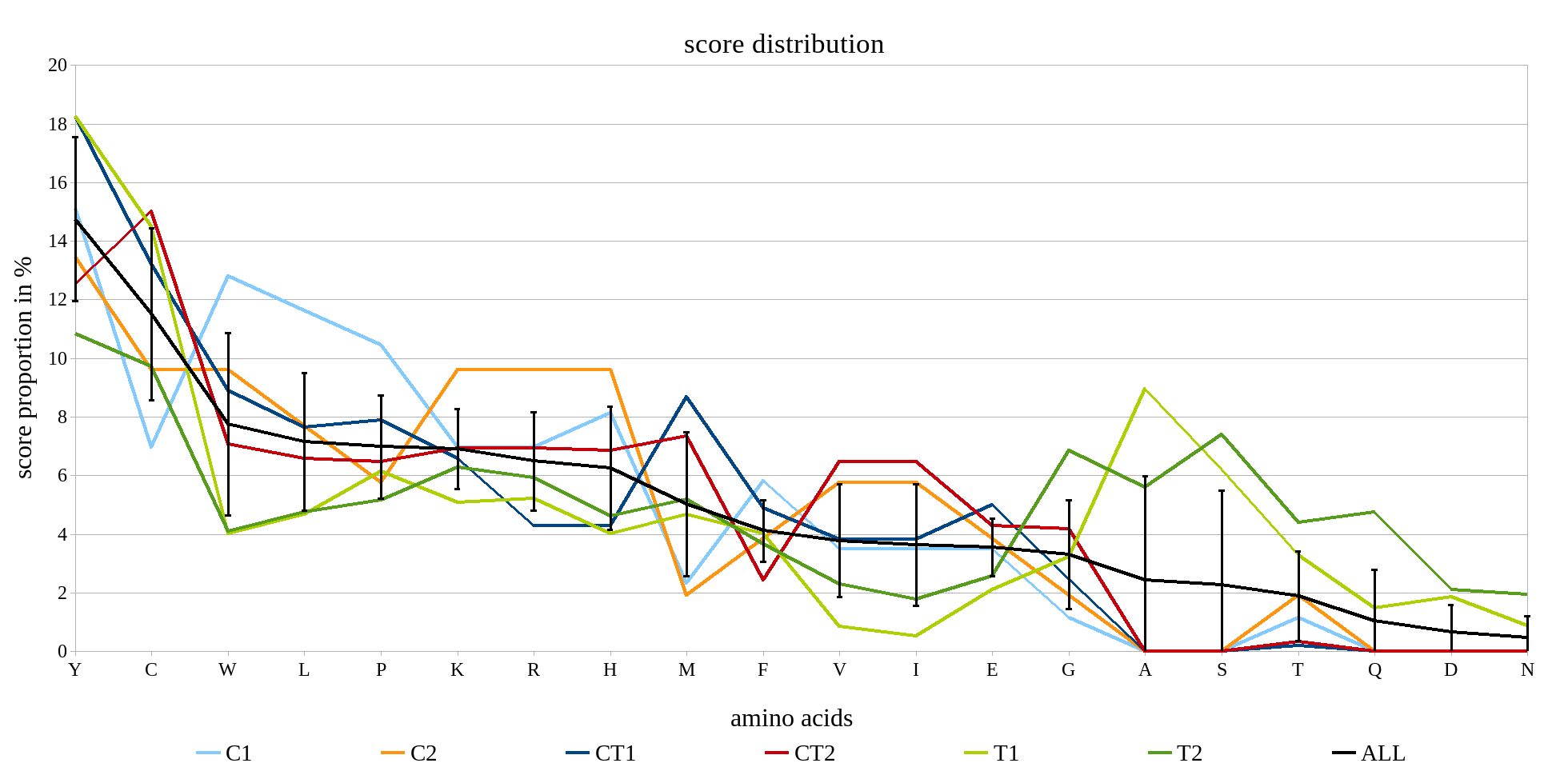
Protein damage (partly followed by protein aggregation) plays a significant role in aging, cancer, and in neurodegenerative and other diseases. It is known that the proteinogenic amino acids differ in their susceptibility to non-enzymatic modification, such as hydroxylation, peroxidation, chlorination, etc. (Fichtner et al., 2020)
More graphics: damage.stark-jena.de
| Amino acid | Modification | Höhn et al. 2013 | Davies 2004 |
Stadtman and Levine 2003 | Berlett and Stadtman 1997 | Key amino acids | Key Modification |
| G (Glycine) | Aminomalonic acid | X | aliphatic | Carbonylation | |||
| I (Isoleucine) | Carbonyls | X | sulfuric | Peroxidation | |||
| Hydroperoxides (unstable) | X | alkalic | Hydroxylation | ||||
| Alcohols | X | acidic | Sulfur oxidation | ||||
| L (Leucine) | A-ketoisocaproic acid | X | aromatic | Nitration | |||
| Carbonyls | X | polar | Chlorination | ||||
| Hydroperoxides (unstable) | X | Bromination | |||||
| Alcohols | X | Dimerization | |||||
| 3-Hydroxyleucine | X | X | Other | ||||
| 4-Hydroxyleucine | X | X | |||||
| 5-Hydroxyleucine | X | X | |||||
| Isovaleric acid | X | ||||||
| Isovaleraldehyde | X | ||||||
| Isovaleraldehyde oxime | X | ||||||
| P (Proline) | Carbonyls | X | |||||
| Glutamic-semialdehyde | X | X | |||||
| Pyroglutamic acid | X | X | |||||
| 2-Pyrrolidone | X | X | |||||
| Hydroperoxides (unstable) | X | ||||||
| Alcohols | X | ||||||
| 5-Hydroxy-2-aminovaleric acid | X | ||||||
| 4-Hydroxyproline | X | X | |||||
| 5-Hydroxyproline | X | X | |||||
| V (Valine) | Carbonyls | X | |||||
| Hydroxyperoxides (unstable) | X | ||||||
| Alcohols | X | ||||||
| C (Cysteine) | Sulfenic acid | X | |||||
| Sulfonamides | X | ||||||
| Cysteic acid | X | X | |||||
| Sulfenyl chloride (unstable; from HOCl) | X | ||||||
| Cystin (disulfid; -S-S- bond) | X | X | X | ||||
| Thiyl radicals | X | ||||||
| M (Methionine) | Methionine sulfoxide | X | X | X | |||
| Methionine sulfone | X | X | X | ||||
| H (Histidine) | Carbonyls (from O2) | X | |||||
| 2-Oxohistidine | X | X | X | X | |||
| Hydroperoxides (unstable; from O2) | X | ||||||
| Alcohols (from O2) | X | ||||||
| Chlorinated materials (unstable; from HOCl) | X | ||||||
| Aspartic acid | X | X | |||||
| Asparagine | X | X | |||||
| L (Lysine) | Carbonyls | X | |||||
| 2-Amino-adipic-semialdehyde | X | X | |||||
| Hydroperoxides (unstable) | X | ||||||
| Alcohols | X | ||||||
| Chloramines (unstable; from HOCl) | X | ||||||
| Bromamines (unstable; from HOBr) | X | ||||||
| R (Arginine) | Carbonyls | X | |||||
| Glutamic-semialdehyde | X | X | |||||
| Hydroperoxides (unstable) | X | ||||||
| 5-Hydroxy-2-aminovaleric acid | X | ||||||
| Chloramines (unstable; from HOCl) | X | ||||||
| Bromamines (unstable; from HOBr) | X | ||||||
| E (Glutamyl) | Hydroperoxides (unstable) | X | |||||
| Oxalic acid | X | ||||||
| Pyruvic acid | X | ||||||
| F (Phenylalanine) | 2,3-Dihydroxyphenylalanine | X | |||||
| 2-Hydroxyphenylalanine (ortho-tyrosine) | X | X | X | ||||
| 3-Hydroxyphenylalanine (meta-tyrosine) | X | X | X | ||||
| 4-Hydroxyphenylalanine (tyrosine) | X | X | |||||
| Nitrophenylalanine | X | ||||||
| W (Typtophan) | Alcohols and cyclized products (from O2) | X | |||||
| Hydroperoxides (unstable; from O2) | X | ||||||
| 2-Hydroxytryptophan | X | X | |||||
| 4-Hydroxytryptophan | X | X | |||||
| 5-Hydroxytryptophan | X | X | X | ||||
| 6-Hydroxytryptophan | X | X | |||||
| 7-Hydroxytryptophan | X | X | X | ||||
| Nitrotryptophan | X | X | |||||
| 3-Hydroxykynurinine | X | ||||||
| N-Formylkynurenine | X | X | X | ||||
| Kynurenine | X | X | X | ||||
| Y (Tyrosine) | Hydroperoxides (unstable; from O2) | X | |||||
| Alcohols (from O2) | X | ||||||
| 3,4-Dihydroxyphenylalanine (DOPA – unstable) | X | X | X | ||||
| 3-Nitrotyrosine | X | X | |||||
| 3-Chlorotyrosine, | X | X | |||||
| 3,5-Dichlorotyrosine | X | X | |||||
| 3-Bromotyrosine | X | X | |||||
| 3,5-Dibromotyrosine | X | X | |||||
| Dityrosine (carbon-carbon dimer, carbon-oxygen dimer, and higher species) |
X | ||||||
| Tyrosine-tyrosine cross-linkages | X | ||||||
| Tyr-O-Tyr | X | ||||||
| Cross-linked nitrotyrosine | X | ||||||
| Cyclized products (from O2) | X | ||||||
| T (Threonine) | 2-Amino-3-keto butyric acid | X | X |
Colorcoder for Fichtner (2019): Download (ZIP)
Chimera: color :ala #FFAAAA; color :cys #FF0000; color :asp #FFEEEE; color :glu #FFDDDD; color :phe #FFEEEE; color :gly #FFEEEE; color :his #FFDDDD; color :iso #FFEEEE; color :lys #FFAAAA; color :leu #FFEEEE; color :met #FFBBBB; color :asn #FFEEEE; color :pro #FFCCCC; color :gln #FFDDDD; color :arg #FFBBBB; color :ser #FFCCCC; color :thr #FFCCCC; color :val #FFEEEE; color :trp #FFDDDD; color :tyr #FF0000;
References:
Fichtner M, Schuster S & Stark H (2021) Influence of spatial structure on protein damage susceptibility: a bioinformatics approach. Sci Rep 11, 4938. DOI:10.1038/s41598-021-84061-8 / EISSN:2045-2322
Fichtner M, Schuster S & Stark H (2021) Data for: Influence of spatial structure on protein damage susceptibility—A bioinformatics approach. Mendeley Data, Licence: CC BY 4.0. DOI:10.17632/jkmbpfgp4k.1
Fichtner M, Schuster S & Stark H (2020) Determination of scoring functions for protein damage susceptibility. Biosystems 187: 104035. DOI:10.1016/J.BIOSYSTEMS.2019.104035 / ISSN:0303-2647
Fichtner M, Schuster S & Stark H (2019) Data for: Determination of scoring functions for protein damage susceptibility. Mendeley Data, Licence: CC BY 4.0. DOI:10.17632/b2cbxsnvcx.1