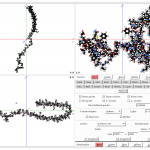
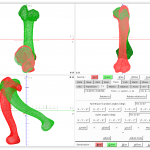
Cloud2 – GUI
Cloud2 is a powerful tool for processing different types of data. Cloud (to) can transform, process, and evaluate data such as point clouds, vector fields, space curves, and meshes. In addition, it has a powerful renderer that allows stunning graphics (see slideshow).
Version 20.12.18

This program is owned by Heiko Stark. It can be used free of charge for non-commercial use. For the common citations in publications, please use the following entry “Cloud2 software (Heiko Stark, Jena, Germany, URL: https://starkrats.de)”.
▸ Download
An installer for Windows and Linux can be found here: Software
cloud2 64bit AVX (Windows & Wine/Darwine)
cloud2 64bit (Windows & Wine/Darwine)
cloud2 32bit (Windows, ReactOS & Wine/Darwine)cloud2 64bit AVX (MacOS X with Quartz/GTK2 & Darling)
cloud2 64bit (MacOS X with Quartz/GTK2 & Darling)
cloud2 32bit (MacOS X with Quartz/GTK2 & Darling)cloud2 64bit AVX (Linux with GTK2)
cloud2 64bit (Linux with GTK2)
cloud2 32bit (Linux with GTK2)
cloud2 aarch64 (Linux with GTK2)
cloud2 arm (Linux with GTK2)cloud2 64bit AVX (FreeBSD with GTK2)
cloud2 64bit (FreeBSD with GTK2)
cloud2 32bit (FreeBSD with GTK2)cloud2 64bit AVX (Solaris with GTK2)
cloud2 64bit (Solaris with GTK2)
cloud2 32bit (Solaris with GTK2)cloud2 64bit AVX (Dragonfly with GTK2)
cloud2 64bit (Dragonfly with GTK2)
cloud2 32bit (Dragonfly with GTK2)
Note: Intel 32bit runs also on 64bit and 64bit AVX systems, but only with 4 GB memory!
Note: Intel 64bit runs also on 64bit AVX systems, but without Advanced Vector Extensions!
Examples
▸ A surface with a hole
// Make a surface with a hole
set.red
set.ref.red
formula.random.point.plane -10 1 10 <0, 0, 0> <0, 0, 1> -10 1 10
bool.inside.circle
bool.erase.cursor.point <0, 0, 0, 0> 4 100
set.ref.green
mesh.clean.surface 0.1
// end
▸ Create a video
// Create a video
for 360
number #= -5 for.count
filename := "image-" number ".ppm"
screenshot.yz filename <1000, 1000>
transform.rotate <0, 0, 1>
end
// end
▸ Norm lines of a graph
// Norm lines of a graph
for object.length
set.red
set.ref.green
bool.move.line
unset.red
set.green
unset.blue
transform.norm.y 0 100
set.green
set.ref.blue
bool.move.line
end
// end
▸ Calculate the mean trajectory of several trajectories.
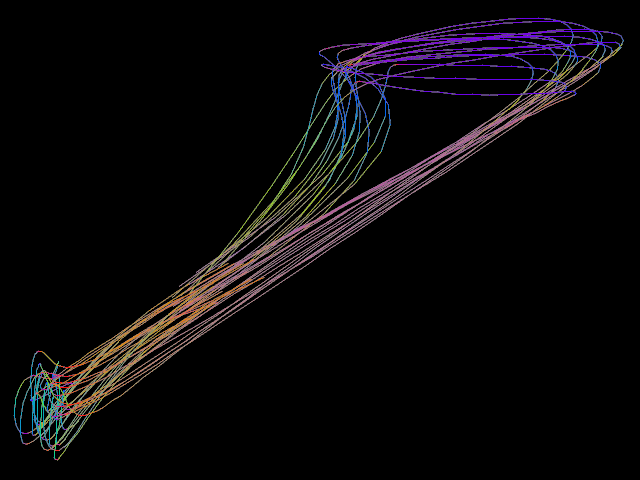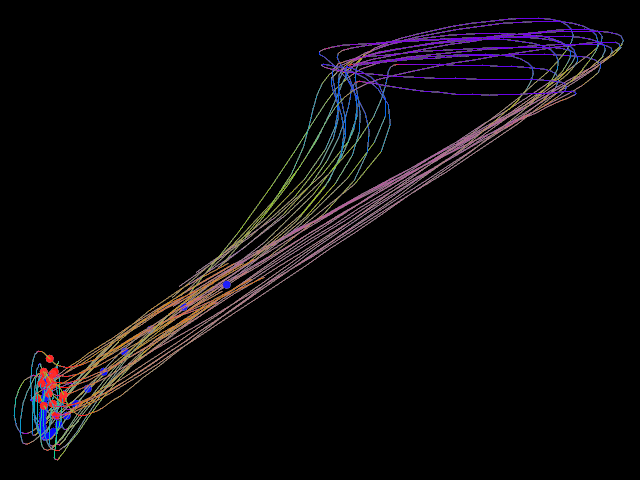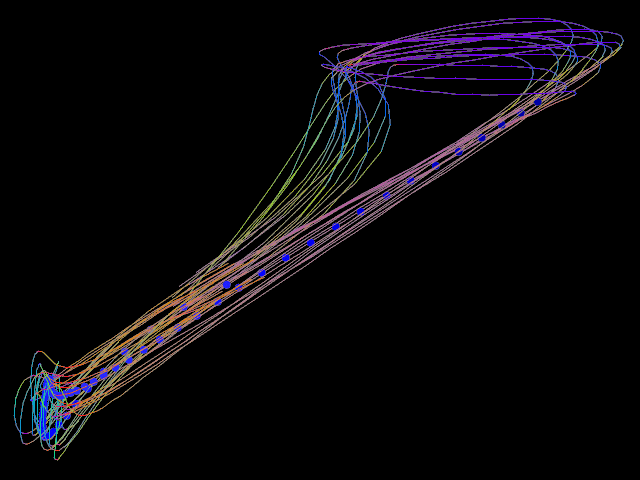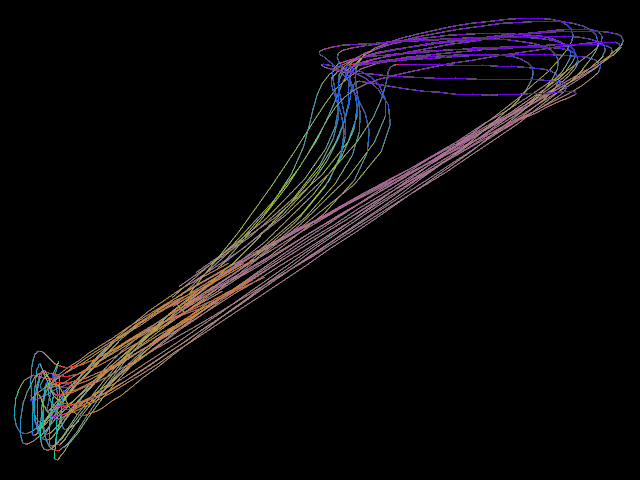
counter := 0 procedure shot number #= -5 counter filename := "/home/tiger/Work/Projekt-Beagle/Modell/Data/2018 04 24/" "image-" number ".png" screenshot.xy filename <640; 480> counter += counter 1 end set.purple unset.cyan set.yellow set.blue set.green set.red new.all set.cyan set.ref.purple bool.copy.layer shot for 1 100 set.cyan set.ref.red calculate.begin.lines.move shot set.red edit.objects.to.one.object calculate.mean // sd shot set.ref.blue bool.move.point shot end set.blue edit.objects.to.one.object edit.objects.to.one.line edit.objects.to.loops edit.smooth.lines 3 5 edit.smooth.lines 3 5 edit.smooth.lines 3 5 shot // saved on date 3.5.2018 | time 14:11:17:43 h
▸ References
Stark H, Fischer MS, Hunt A, Young F, Quinn R, Andrada E (2021) A three-dimensional musculoskeletal model of the dog. Sci Rep. 11(1), 11335. DOI: 10.1038/s41598-021-90058-0 / EISSN:2045-2322
Fichtner M, Schuster S & Stark H (2021) Influence of spatial structure on protein damage susceptibility: a bioinformatics approach. Sci Rep 11, 4938. DOI:10.1038/s41598-021-84061-8 / EISSN:2045-2322
Sartori J, et al. (2021) Gaining Insight into the Deformation of Achilles Tendon Entheses in Mice. Adv. Eng. Mater. 2100085. DOI:10.1002/adem.202100085
Sartori J, Stark H (2021) Tracking tendon fibers to their insertion – a 3D analysis of the Achilles tendon enthesis in mice. Acta Biomaterialia 120: 146-155. DOI:10.1016/j.actbio.2020.05.001 / ISSN:1742-7061
Nyakatura JA, Baumgarten R, Baum D, Stark H, Youlatos D (2019) Muscle internal structure revealed by contrast-enhanced µCT and fibre recognition: The hindlimb extensors of an arboreal and a fossorial squirrel. Mammalian Biology 99: 71-80. DOI:10.1016/j.mambio.2019.10.007 / ISSN:1616-5047
Rosin S & Nyakatura JA (2017). Hind limb extensor muscle architecture reflects locomotor specialisations of a jumping and a striding quadrupedal caviomorph rodent. Zoomorphology 136: 267. DOI:10.1007/s00435-017-0349-8
Kupczik K, Stark H, Mundry R, Neininger F, Heidlauf T, Röhrle O (2015). Reconstruction of muscle fascicle architecture from iodine-enhanced microCT images: a combined texture mapping and streamline approach. Journal of Theoretical Biology 382 (7): 34–43. DOI:10.1016/j.jtbi.2015.06.034 / ISSN:0022-5193
Nyakatura JA, Stark H (2015). Aberrant back muscle function correlates with intramuscular architecture of dorsovertebral muscles in two-toed sloths. Mammalian Biology – Zeitschrift Für Säugetierkunde 80 (2): 114–121. DOI:10.1016/j.mambio.2015.01.002 / ISSN:1616-5047
Stark H, Fröber R, Schilling N (2013) Intramuscular architecture of the autochthonous back muscles in humans. Journal of Anatomy 222 (2): 214–222. DOI:10.1111/joa.12005 / ISSN:0021-8782
Stark H, Schilling N (2010) A novel method of studying fascicle architecture in relaxed and contracted muscles. Journal of Biomechanics 43 (15): 2897-2903. DOI:10.1016/j.jbiomech.2010.07.031 / ISSN:0021-9220
▸ Screenshots
If you want to send me a bug report or have some suggestions about what future versions of ‘cloud2’ should support, you can contact me either by email (bugs[@]starkrats[dot]de).